Bioinformatics Training Platform (BTP) User Guide
Introduction
Monash eResearch Centre have been partnering with Bioplatforms Australia (BPA) and CSIRO for delivering hands-on bioinformatics workshops to Australian researchers. This partnership started back in 2012 when the first next-generation sequencing workshop was delivered at Monash University. The Bioinformatics Training Platform (BTP) has been developed as a broadly accessible solution for delivering these workshops using cloud computing infrastructures and addressing the increasing demand for training workshops in Australia.
The standard NGS workshop delivered by BPA and CSIRO is composed of a number of training modules. Each training module is composed of the following components: analysis tools, datasets and training materials. The analysis tools are essential to a hands-on bioinformatics workshop as it teaches scientists how such tools are used to interrogate and interpret results. Most of the analysis tools used by the BTP are open source and easily accessible on various platforms. The BTP maintains these analysis tools on BPA-CSIRO GitHub Project. To support the hands-on bioinformatics workshop, analysis and reference datasets are used together for analysis. Datasets are stored and managed on the NeCTAR object storage service. The training materials (presentations and handouts) are also developed and maintained as part of the BTP. A high level overview of the BTP architecture is show below:
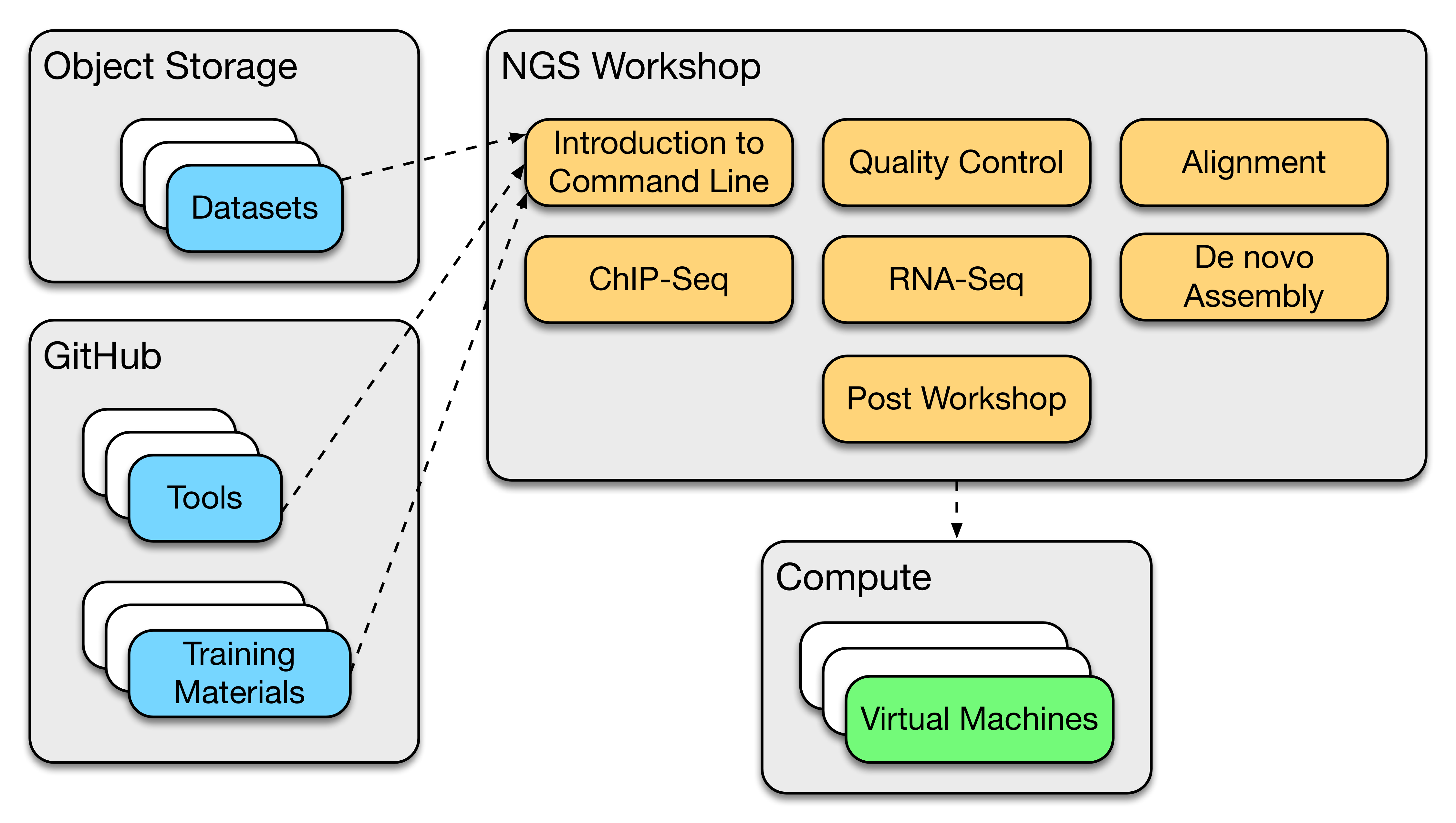
A workshop is composed of a number of training modules. Each training module has an associated analysis tools and datasets metadata. These metadata are used by the orchestration system for automation of tools installation during BTP image creation and datasets syncing during BTP instance deployments.
Components
The BTP as a solution is composed of a number of components: Analysis Tools, Datasets and Training Materials. These components are encapsulated on each of the training modules. A brief summary of each one are included below.
Analysis Tools
Analysis tools are essential for delivering hands-on training workshops. The BTP teaches trainees how these tools can be used to help interrogate and interpret their analysis results. Most of these analysis tools are available as stand alone executables while others have to be built from source. The most common analysis tools used and packaged in the BTP are listed in the table below:
| Tools | Function |
|---|---|
| AMOS Hawkeye | Genome data visualization |
| BEDTools | Genome data manipulation |
| BLAT | Sequence location lookup in the genome |
| Bowtie | Read Alignment |
| CummeRbund | RNA-Seq analysis using R |
| Cufflinks | RNA-Seq analysis |
| DESeq2 | Differential gene expression analysis using R |
| edgeR | Empirical gene expression analysis using R |
| FastQC | FastQC |
| FASTX | Toolkit for short reads preprocessing |
| IGV | Interactive exploration of genomic data |
| igvtools | Preprocessing of data before loading to IGV |
| MACS | ChIP-Seq analysis |
| MUMmer | Rapid genome alignment, a dependency for AMOS |
| PeakAnalyzer | Multi-peak data analysis |
| Picard | Sequence data analysis |
| SAMtools | For manipulating alignments in the SAM format |
| Skewer | Adapter trimmer for paired-end reads |
These analysis tools are packaged and installed into the BTP Image upon creation. This process is described in detail in the [BTP Workflows][#btp-workflows] section below.
Packaging of Analysis Tools
To make maintenance and deployment of the BTP easier, most of the analysis tools included in the BTP are packaged as stand-alone *.deb installers, installable on Ubuntu-based operating systems. fpm-cookery is used to create the *.deb installers using recipes. The current BTP Images are based on Ubuntu Precise LTS which is supported until 2017. The BTP maintains a set of recipes on GitHub which is also integrated with Travis CI for continuous integration. Whenever a tool recipe is updated or new recipe is added into the repository, Travis CI ensures that the recipes builds correctly.
The .deb installers can also be manually created on a client machine. This step is not mandatory for creating the BTP images and deployment of BTP instances. One can clone the BTP Tools repository, and using fpm-cookery, create the .deb installers.
Clone the BTP Tools repository:
git clone https://github.com/BPA-CSIRO-Workshops/btp-tools.git
The .deb installer for a particular analysis tool can then be built using fpm-cookery:
cd btp-tools/bwa
fpm-cookery .
fpm-cookery will read the instructions in the recipe and build the .deb installer. In the example above, fpm-cookery will get the source code for bwa from GitHub, then will proceed with the standard configuration, compilation and building of the tool. fpm-cookery will place the resulting .deb installer inside the pkg subdirectory:
ls pkg/
bwa_0.7.12-0_amd64.deb
Datasets
Datasets used by the BTP are stored on NeCTAR Object Storage (Swift). Each training modules has a corresponding publicly readable container where its datasets are accessible. The containers for the NGS training modules are listed in the table below:
| Module | Container |
|---|---|
| Introduction to CLI | NGSDataCommandLine |
| Quality Control | NGSDataQC |
| Alignment | NGSDataChIPSeq |
| ChIP-Seq | NGSDataChIPSeq |
| RNA-Seq | NGSDataRNASeq |
| De novo Assembly | NGSDataDeNovo |
The complete container URL are used to pull down the datasets into the BTP instances. The URLs are defined on the datasets metadata file, which is used by Puppet to pull down the datasets into the BTP instances upon deployment.
Training Materials
BTP Workflows
Creating the BTP Image
Prerequisites for Creating BTP Images
- git
- Packer
- OpenStack Glance Command Line Client
- NeCTAR OpenStack Credentials
- internet access
The BTP uses the Packer tool for creating new virtual machine images compatible with NeCTAR Research Cloud.
Get Workshop Repository
A collection of Packer recipes are included as part of the orchestration module inside the BPA-CSIRO Workshop NGS Repository. This repository must be cloned together with the training submodules on the client machine where the BTP image will be created:
git clone --recurse-submodules https://github.com/BPA-CSIRO-Workshops/btp-workshop-ngs.git
Orchestration
Once the workshop repository and the training submodules have been cloned, the local copy will have the following contents structure:
├── 010_trainers
├── 015_preamble
├── 050_ngs-qc
├── 060_alignment
├── 070_chip-seq
├── 080_rna-seq
├── 090_velvet
├── 905_post-workshop
├── Makefile
├── README.md
├── developers
├── licences
├── orchestration
├── style
└── template.tex
The numberings (e.g 010_trainers, 015_preamble) are used by the build system to order by which the training modules are ordered in the training handouts. Each training module has an associated handouts subdirectory where the LaTeX file for that module is located. Going inside the orchestration/packer subdirectory will show the available Packer recipes including the one compatible with the NeCTAR Research Cloud, btp-qemu.json. The available Packer recipes for the BTP are:
├── btp-aws.json
├── btp-qemu.json
├── btp-virtualbox.json
├── btp-vmware.json
Launch Packer
Before launching Packer, the content of the recipe file btp-qemu.json can be viewed. Descriptions for each recipe section are highlighted below.
The image creation process can then be started by feeding the recipe into Packer. This assumes that the Packer is already installed on the client (build) machine. Packer can then be launched, with the recipe filename passed as a command line argument:
packer build btp-qemu.json
Packer will start the image creation process and the first thing this will do is to download the latest available alternate Ubuntu ISO from the internet. So the client machine must have access to the internet to download the ISO file.
Upload Image
Deploying BTP Instances
Prerequisites for Deploying BTP Instances
- NeCTAR OpenStack Credentials
- OpenStack Nova Command Line Client
- NoMachine (NX) Client
Using the BTP Instances for Training
Was this article helpful?
That’s Great!
Thank you for your feedback
Sorry! We couldn't be helpful
Thank you for your feedback
Feedback sent
We appreciate your effort and will try to fix the article